Advancing data-driven drug discovery with interactive biomedical evidence
Most new drug development programs start with foundational pre-discovery research that includes in-depth searches of the literature as well as public data repositories and other biomedical information resources. The process of assembling this library of evidence is incredibly time consuming and tedious. Findings are too often stored in separate spreadsheet files with poor version control and infrequent updates.
Aevidence changes all that.
This biological intelligence platform comprises 10 specialized discovery apps for interactively exploring different types of critical evidential data from public and internal data resources. It provides a framework to gather, organize, update, and share the supporting information needed to:
- Investigate potential biological targets, such as receptors, enzymes, proteins, molecular structure, genes, and other features that may drive mechanisms underlying a disease of interest
- Leverage competitive intel on companies, approved medicines and treatments, ongoing clinical trials, and market potential for novel medicines and therapeutic modalities.
- Inform experimental design and assess results of early discovery efforts
When you pair the Aevidence Platform and visyn Knowledgebase with Ordino, you elevate and optimize your discovery data analysis workflows with immediate access to a wealth of scientific evidence and public data – all in one turbocharged visual analytics suite.
Perform informed visual analysis with built-in biological knowledge
1.
Search for Entities
Gain access to an exhaustive biological knowledgebase that provides insights and supporting evidence to help you validate discovery findings. Whether you’re searching for genes, diseases, compounds, or companies, Aevidence enables you to quickly retrieve the detailed information you want about any combination of entities–without leaving your datavisyn workspace.
2.
Retrieve Query Results Instantly
Get invaluable insights with easy-to-build queries. Aevidence enables you to explore different types of biological evidence using a suite of specialized discovery apps, each with an intuitive design that best fits the type of data you’re mining, whether genetic sequences, protein structures, or known drug interactions. Get a comprehensive overview of all your evidence in realtime, and save searches for every entity and their combinations, so you can retrieve updated results in seconds.
3.
Explore Biomedical Evidence in Context
Drill down and dig deeper to investigate the nuances of your data to identify key takeaways. The visyn Knowlegebase gives you immediate access to a wealth of biological intelligence that helps you quickly answer how and why questions about the role of entities of interest. It includes tools that simplify data uploads and integration, so you can further enrich the knowledgebase with your in-house research data and perform explore evidence tailored to your drug program.
All the Evidence You Need


Dive deep into disease biology
Rely on Aevidence to mine the visyn Knowledgebase of regularly updated biomedical intelligence to strengthen your understanding of disease pathophysiology and rigorously assess potential targets and therapeutic modalities in the context of your disease of interest — all in one versatile platform.

Leverage specialized apps and vis tools
Gain insights from a wide range of powerful applications designed for biological data visualization, including species-specific bioactivity data from ChEMBL and GOStar, protein structure bioinformatics from RCSB, and protein sequence identify data for homologs and orthologs in relevant model organisms.

Visually explore entities to extract insights
Use keywords to search for biological entities, and reveal a comprehensive overview of available data. Explore biological data in realtime, using interactive tools designed to help you dig into underlying data for disease genes, expression studies, structural bioinformatics, proteomics, pipelines, and so much more.
Extend our platform with your data
Supplement the visyn Knowledgebase’s extensive collection of preloaded public datasets with your in-house experimental and patient data, then leverage intuitive workflows for extracting even greater insights that will inform go/no go decision making at every stage of preclinical drug discovery and development.
Elevate Your Visual Analyses with Integrated Biomedical Intelligence
Aevidence in Numbers
Aevidence: Interactive, Intuitive, Inspiring
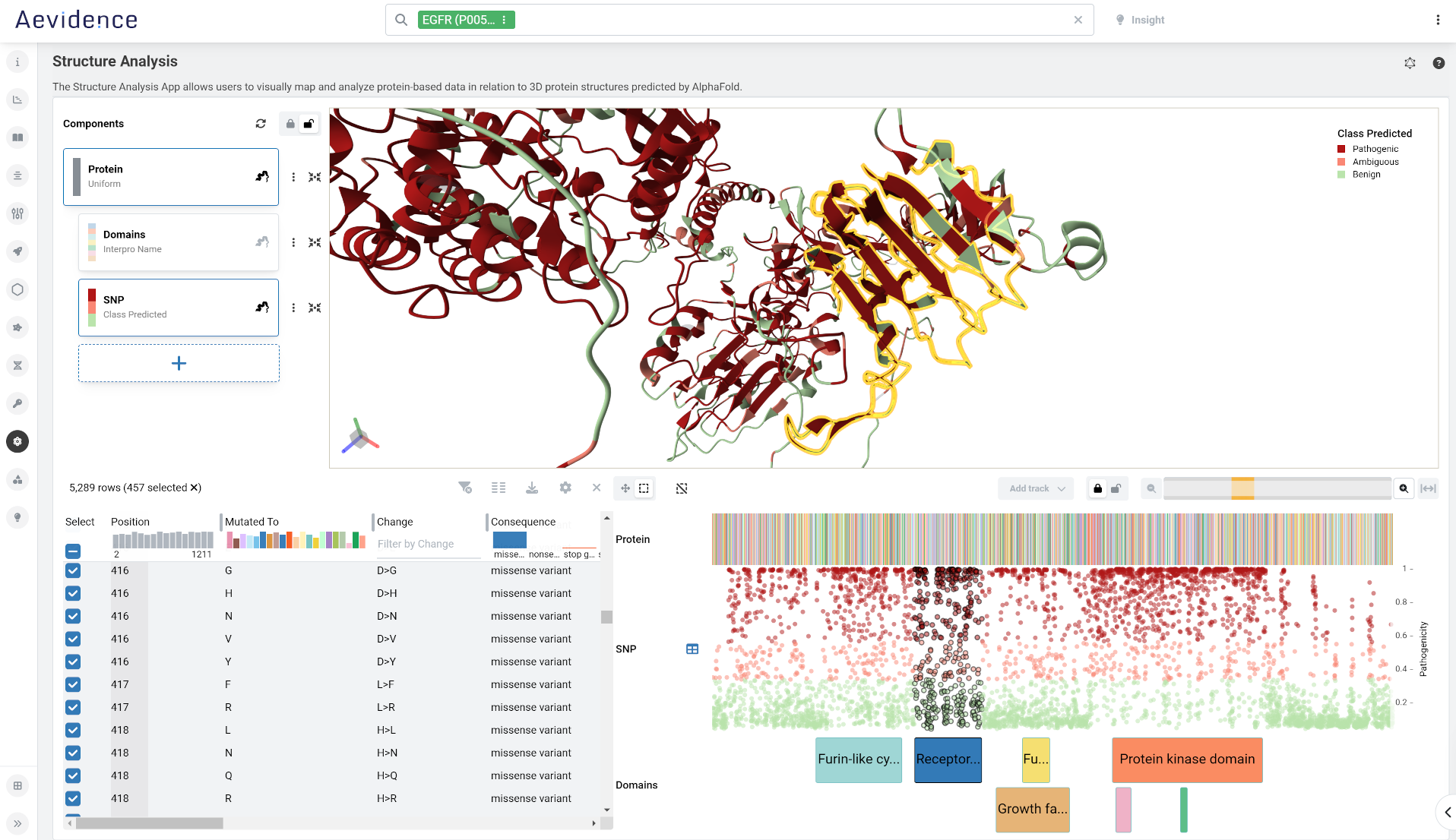
Structural Bioinformatics
Visualize 3D structures and explore underlying sequences of experimentally-determined and computed proteins in the RCSB protein data bank. Understanding the 3D structure of a biological macromolecules is critical for discerning its role in health, disease, and therapeutics.
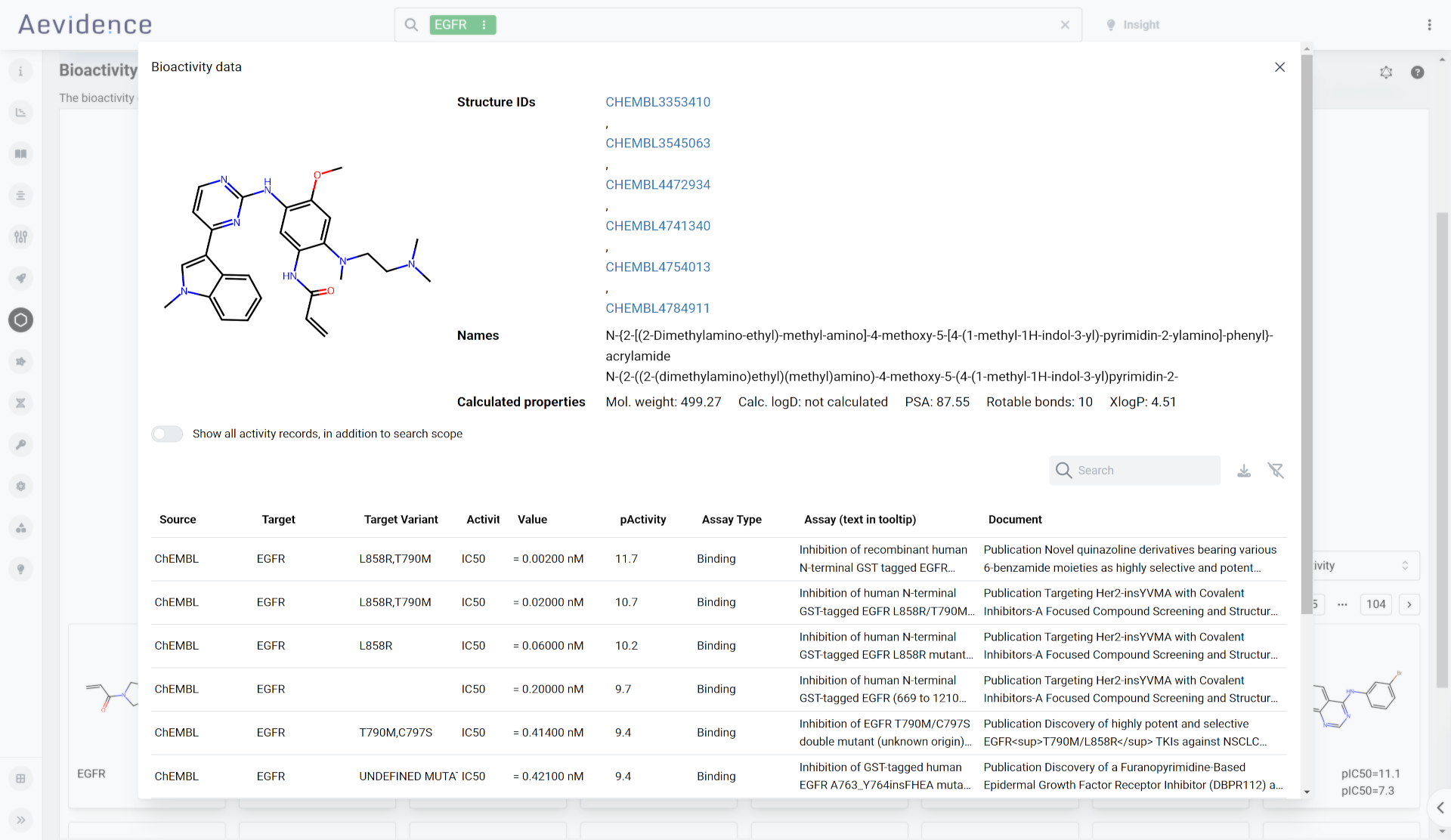
Bioactivity
Investigate comparative human, mouse, and rat data from ChEMBL and PROTAC-DB. Here you can review characteristics of bioactivity molecules with drug-like properties and proteolysis-targeting chimeras (PROTACs). You can also define target and off-target filters as well as structural constraints, such as substructures or reference structures for similarity filtering.
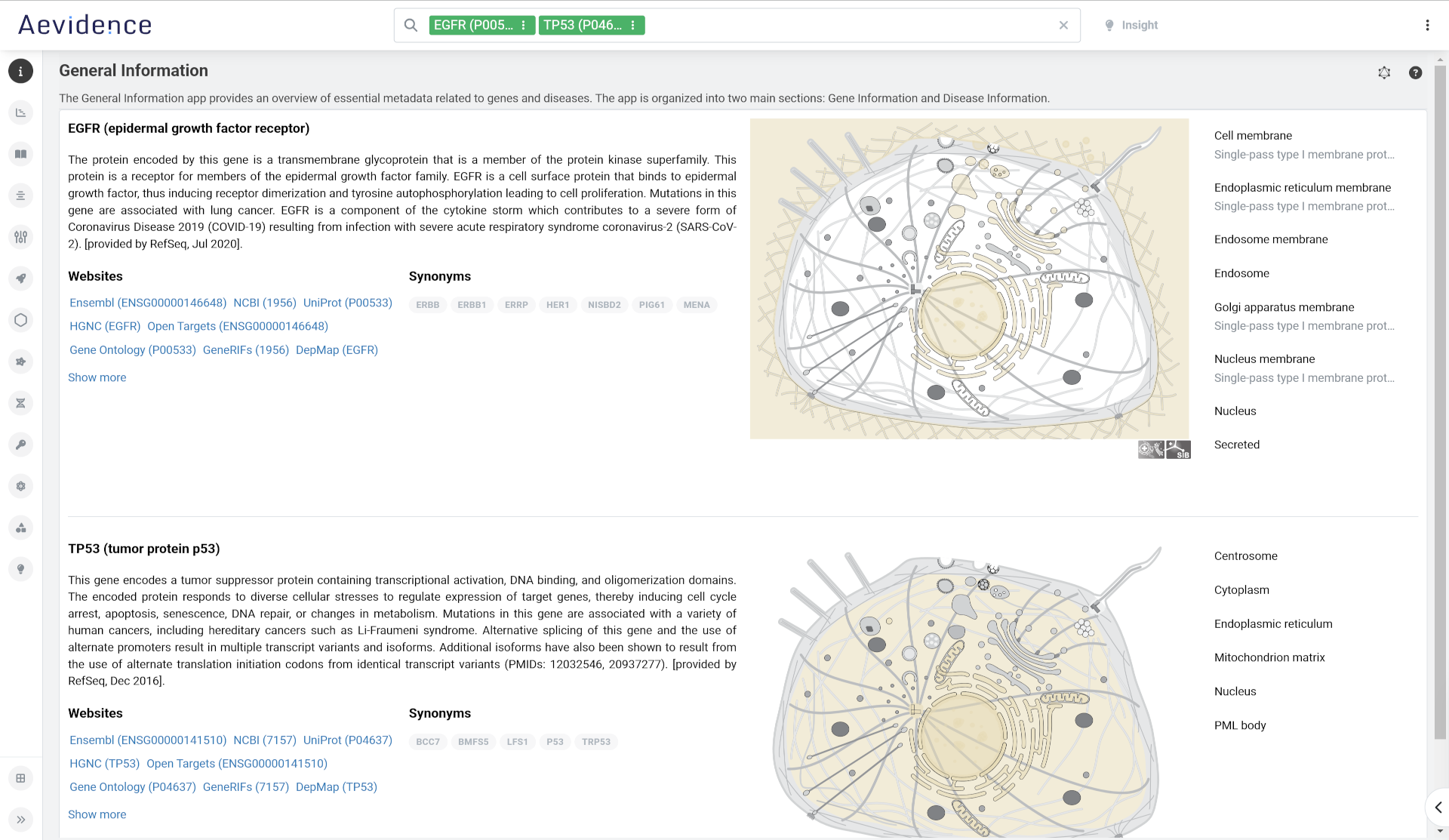
General Information
Retrieve a concise summary of your genes or diseases of interest and their function. For genes, the subcellular location view provides information on known location and topology of mature proteins within the cell and displays where within the cell a protein of interest is expressed. Click on a description to highlight the organelle in the cell diagram.
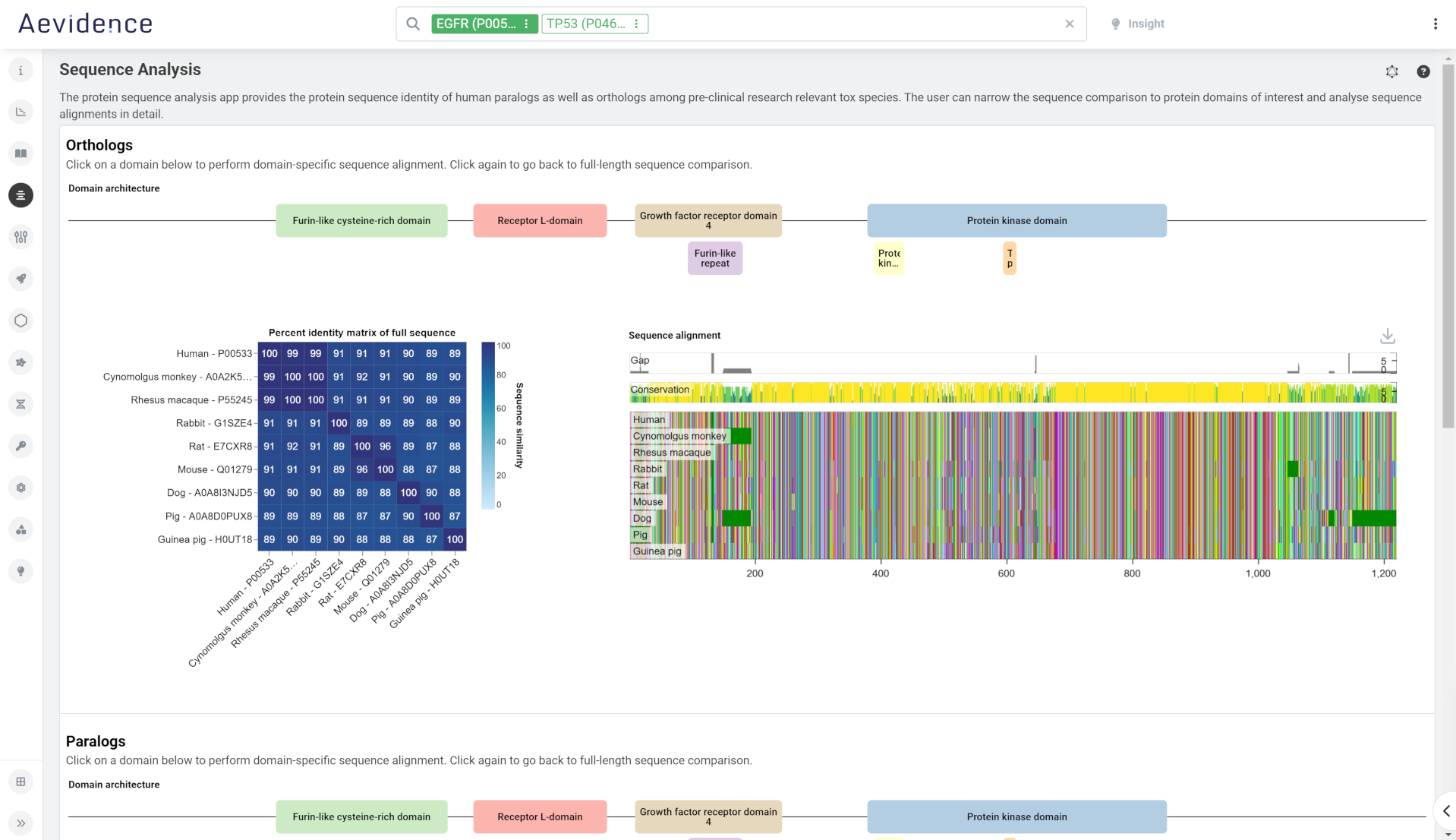
Protein Sequence Analysis
Retrieve protein sequence alignments between human and animal species commonly used in preclinical toxicology studies. Here you can view human gene orthologs for selected species represented in NCBI orthologs and OrthoDB, as well as a table of homologous human proteins. For homologs, you can run a multiple sequence alignment on selected homolog proteins (using sequence data from the UniProt Knowledgebase. You can narrow the sequence comparison to focus on protein domains of interest as you analyze sequence alignments in detail.
More Aevidence Apps
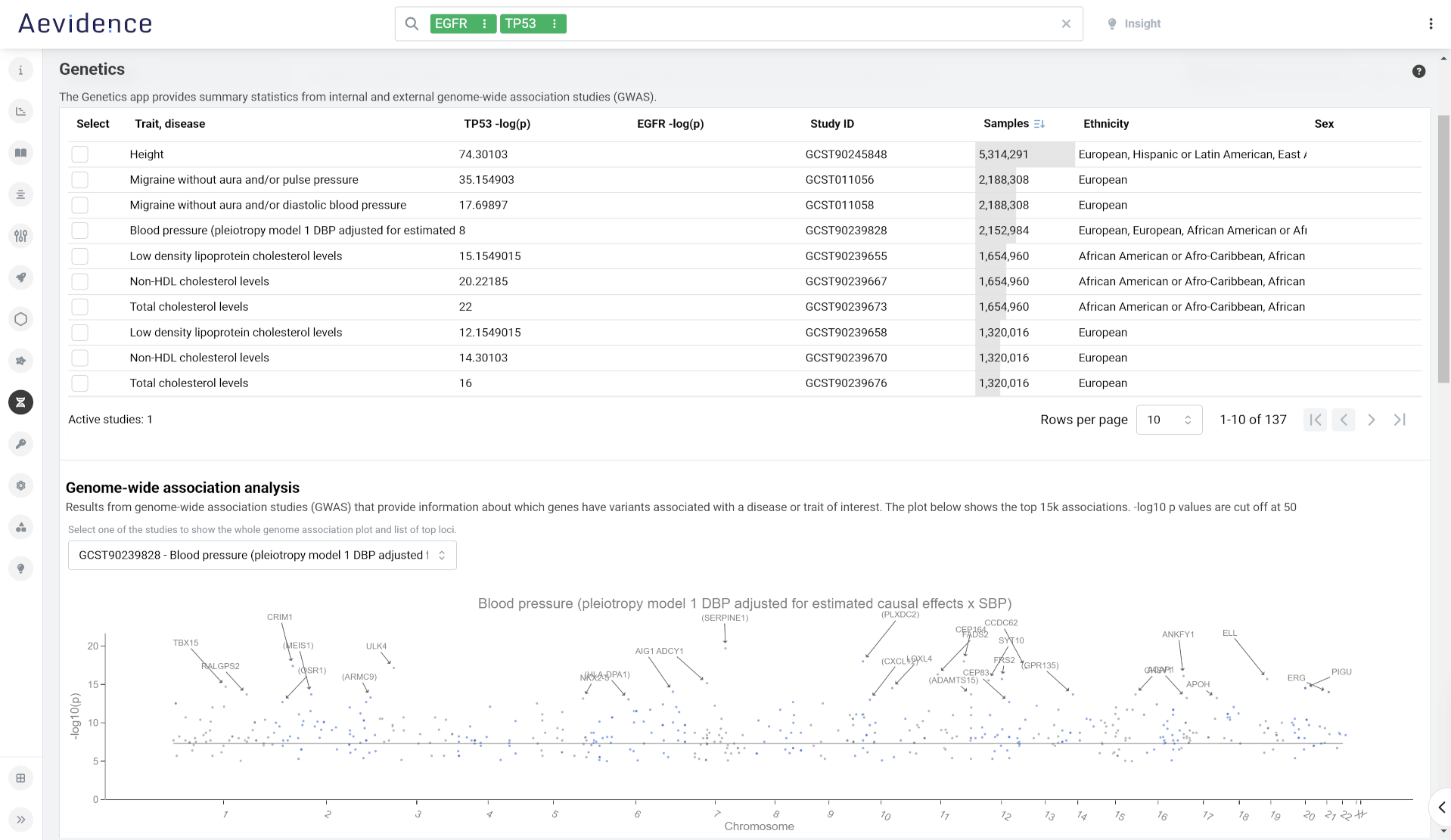
GWAS and PheWAS Studies
Get summary statistics from internal and publically available data from genome-wide association (GWAS) studies and PheWAS phenome-wide association (PheWAS) studies, with links to publications in PubMed. Select a study to review GWAS study information about genes with variants associated with a specific disease or trait, and drill down to see a regional association plot of any trait associations within a given genomic range.
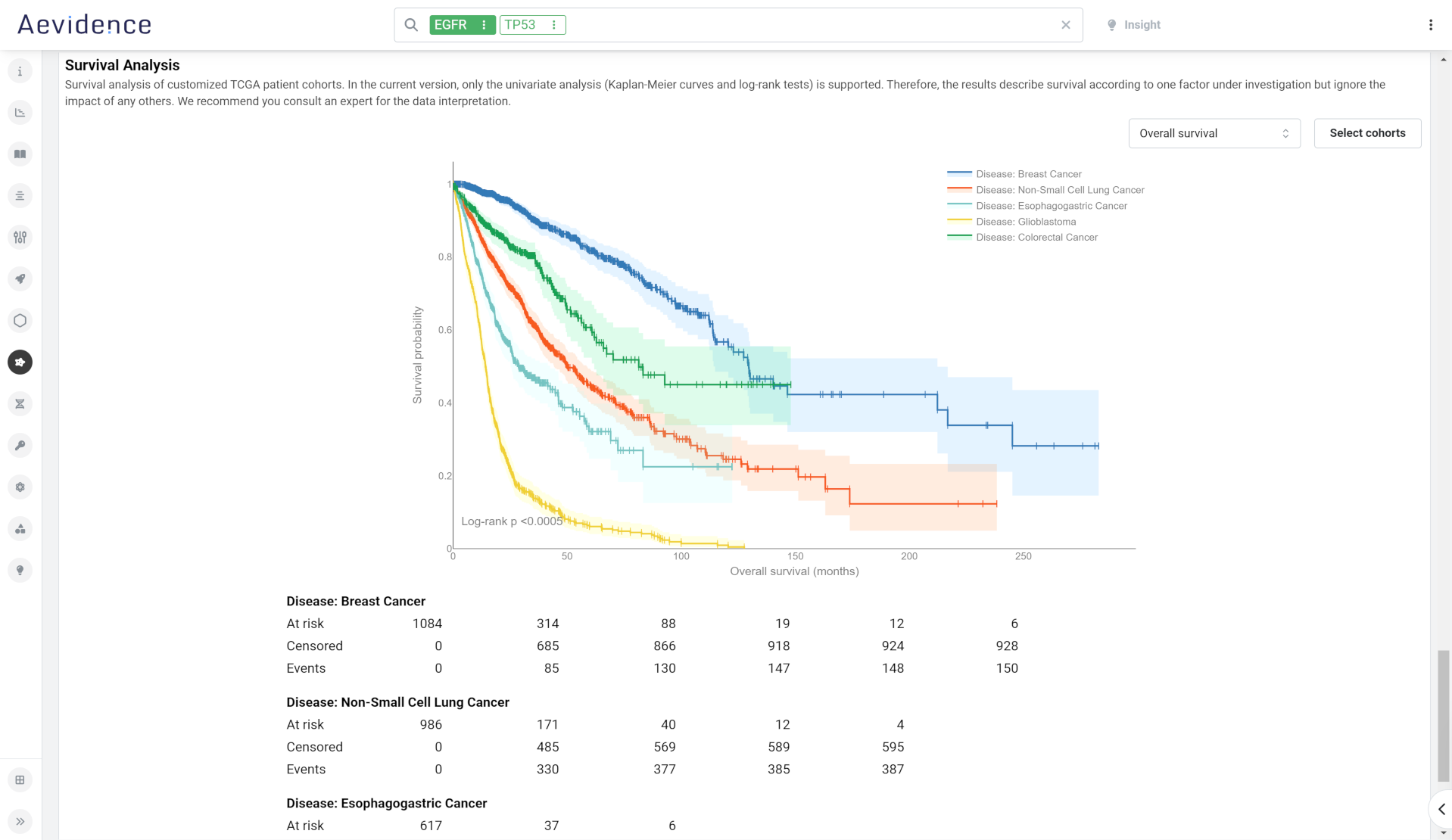
Oncology
Access a visual overview of genomics alterations documented in The Cancer Genome Atlas (TCGA) . For selected genes, this portal presents genomic alteration prevalence by cancer type, gene expression distribution in tumor and normal samples, missense mutation hotspots, DepMap gene effect scores for CRISPR knock-outs across different cell lineages and survival analyses for selected patient cohorts.
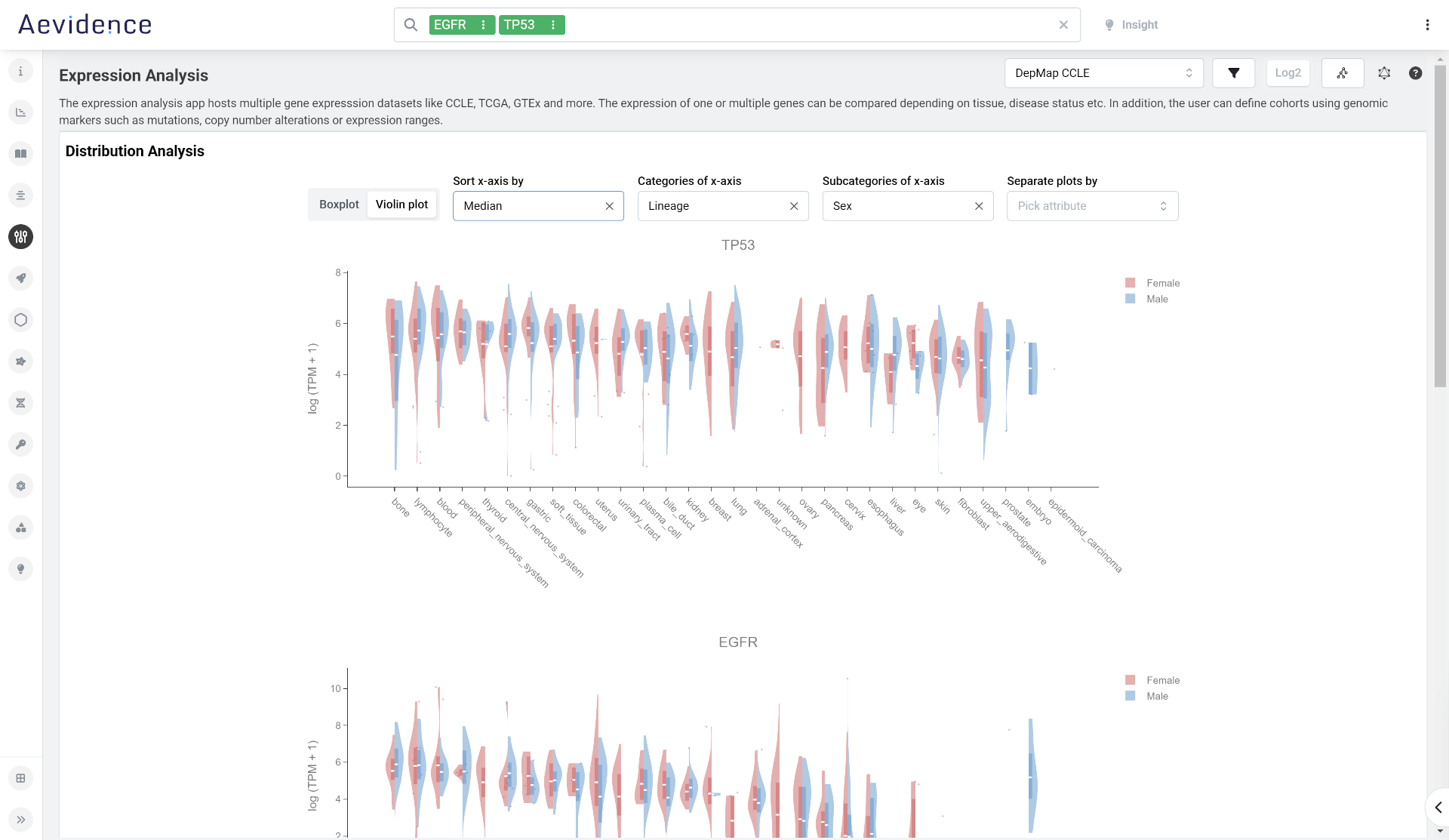
Expression Analysis
Compare expression of one or multiple genes using integrated data from the Genotype-Tissue Expression (GTEx) portal, The Cancer Genome Atlas (TCGA), and Cancer Cell Line Encyclopedia (CCLE). Here, you can visualize expression data in the context of a specific tissue type, disease status, or other factors. You can also define cohorts using genomic markers (e.g., mutations, copy number alterations, expression ranges, etc.).
Develop your Own Aevidence App
Work with us to create new portals tailored to fit your requirements and discovery data landscape.